弊社が販売する試薬は全て研究用です。為替変動等の影響により、ページ内に記載されている料金等の金額は全て参考につき、詳細はお気軽にお問い合わせください。
・各種受託分析トップページへ戻る
・関連製品 ・ サービスへ進む
・お問い合わせ ・ ご注文へ進む
This service / product is supplied by  .
.
弊社が提供するCreative Biolabs社のシーケンシングサービスは研究用です。臨床診断用ではありません。
目次

SuPrecision™次世代シーケンス関連受託サービス
次世代シーケンシング (NGS)は、がん研究等において重要な役割を担っています。バイオロジカはCreative Biolabs社によるNGS関連サービスをフルレンジでカバーするSuPrecision™ プラットフォームを提供しています。このツールは、多面的な疾患に対する我々の理解を大きく向上させ、正確で高感度、ハイスループットかつ費用対効果の高いがん研究を実現します。
ご存知のように個別化医療とは、個人の遺伝的プロファイルを利用して健康管理をカスタマイズするもので、急速に発展しているヘルスケア産業です。次世代シーケンシング (NGS)とコンピューター解析の爆発的な進歩により、がんの予防や早期発見を目的とした評価が可能になりました。同社は、NGS分野における長年の研究開発経験をもとに、先進のSuPrecision™プラットフォームを構築し、がん診断やプレシジョンメディシンを支援する正確かつハイスループットのシーケンシングサービスを提供します。
詳細は以下リンク先のメーカーサイトをご覧ください。
 | がんの全ゲノムシーケンシング (WGS)サービスWGSの開発と技術の低価格化により、ヒト疾患の遺伝的予測因子と関連する治療の提供が加速することが期待されています。WGSは、がんにおけるあらゆる種類のゲノム変化を包括的に調査し、がんゲノムにおけるドライバー変異および変異シグネチャーの全体像をよりよく理解し、そして、これらの未踏のゲノム領域および変異シグネチャーの臨床的意味を明らかにするために使用することができます。従って、WGSは変異の解釈とがん精密医療のための重要な分析プラットフォームです。 |
 | がんの全エキソームシーケンシング (WES)サービスWESは、ヒトの疾病表現型に関連する遺伝的変異の大部分が存在するエクソンの95%以上をカバーしています。WESは、ゲノムの最も関連性の高い部分に焦点を当てることで、研究者がシーケンシングと解析リソースをより効率的に使用できるようにし、一般的および稀な変異の発見と検証を容易にしています。WESは、複数の医学的問題の統一的な診断を探している患者さんに特に適しています。 |
 | がんのターゲットシーケンシングサービスターゲットシークエンシングは、アンプリコンや特定の遺伝子に着目したNGS技術であり、様々な遺伝子疾患の診断に有効なアプローチです。ターゲットシーケンス技術は、がん患者に用いられるプレシジョンメディシンの基礎を提供するものとして、その臨床的有用性が実証されています。 |
 | がんの全トランスクリプトームシーケンシング (WTS)サービスWTSは、ヒトのがんを含む多くの疾患の分子メカニズムを理解するための重要なツールです。同社は、mRNAトランスクリプトームシーケンシング (mRNA-Seq)、マイクロRNAシーケンシング (miRNA-seq)、サーキュラーRNAシーケンシング (circRNA-seq)、ロングノンコーディングRNAシーケンシング (lncRNA-seq)など、一連のがんWTSサービスを提供しています。 |
 | がんの免疫レパートリーシーケンシング (Rep-Seq)サービス近年のハイスループットな免疫レパートリー解析の出現により、B細胞およびT細胞受容体 (BCRおよびTCR)レパートリーの多様性を直接理解することができるようになりました。同社は、単一のサンプルに含まれるTCR (α、β、γ、δ)およびBCR V (D)J配列の同定において豊富な経験を蓄積しています。 |
 | エピジェネティクスサービスエピゲノミクスとは、DNA配列を変えずに遺伝子の活性や発現に影響を与える一連のエピゲノム修飾のことですが、DNAメチル化やヒストン修飾など様々なエピゲノム修飾が、がんの病態と関連することが報告されています。 |
 | 3Dゲノミクスサービスゲノムの3次元配置は、遺伝子の制御にとって極めて重要ですが、ゲノムは複雑かつ動的であり、細胞の種類によってや、細胞の分化や発生の過程で変化することがあります。ゲノムの働きを理解するためには、染色体構造を研究する必要があります。 |
 | がんのメタゲノム解析サービス何千もの微生物種が、正常なヒトのマイクロバイオームを構成していますが、その微生物群は、正常な生理機能や、がんを含む疾患への反応に大きな影響を与える可能性があります。 |
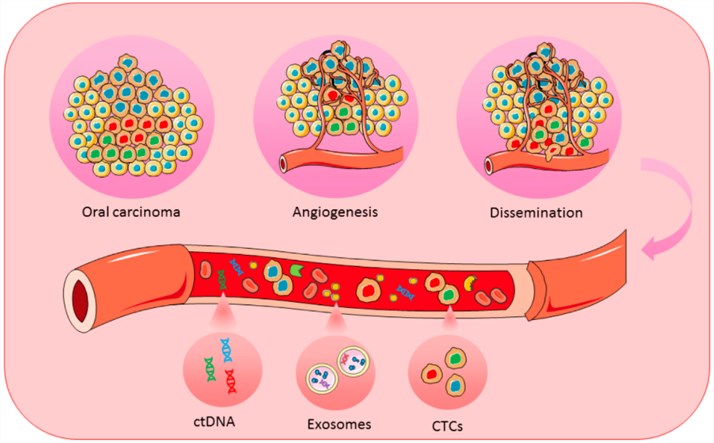 | ctDNAシーケンシングサービスctDNA検査は、リキッドバイオプシーとも呼ばれ、組織生検に代わる低侵襲な診断法として、腫瘍診断や予後予測への応用が期待されています。NGSは、血中の低レベルのctDNAを高感度かつ特異的に検出できる優れた技術を提供します。 |
 | がんの単一細胞シーケンシングサービスシングルセルシーケンシング技術は、単一細胞の遺伝子発現、DNA変異、エピゲノム状態、核構造を解析する画期的なツールであり、発がんや腫瘍進行の分子機構の理解を深めることができます。 |
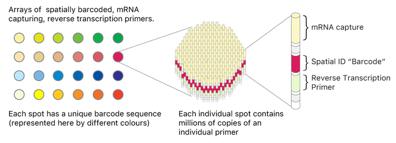 | がんの空間的トランスクリプトームシーケンス空間トランスクリプトームシーケンシング技術は、個々の細胞の遺伝子発現を本来の場所で調べるための強力なツールであり、病理学、免疫学、発生生物学、神経科学、腫瘍学を含む多くの研究分野で利用されています。 |
Creative Biolabs社は、世界をリードする最新のテクノロジープラットフォームと専門的な科学スタッフを備え、がん研究をサポートするために高品質のNGS基盤のサービスとバイオインフォマティクス解析サービスを提供することに専心しています。
詳細な情報および見積もりについては、こちらからお問い合わせください。
