Mechanism | Description | Protocol | Inquiry / Order
A new labeling method to the target protein on the cell surface
– Easy ID™ –
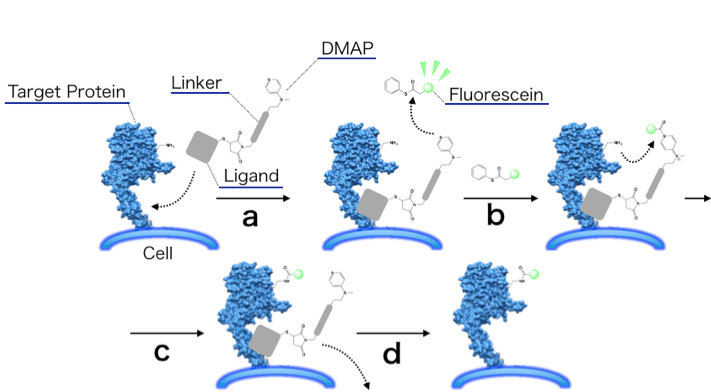
Mechanism of “Easy ID™”
Innovative labeling system by AGD chemistry (Affinity-Guided DMAP chemistry)
This system is the labeling method which can attach ** Acyl donor (thioester derivative – fluorescent dye modified) to the target protein with * DMAP linked to ligand as a catalyst.
Fluorescence-modified small molecules are covalently bonded to the target instead of ligands for receptors, antibodies (VHH, scFv, Fab) to antigens.
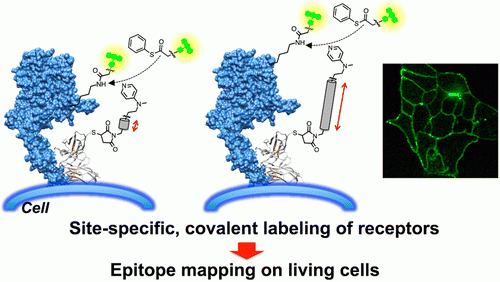
“Analysis of Cell-surface Receptor Dynamics Through Covalent Labeling by Catalyst-tethered Antibody” †
Takahiro Hayashi, Yuki Yasueda, Tomonori Tamura, Yousuke Takaoka, Itaru Hamachi *
J. Am. Chem. Soc., 137, 5372-5380 (2015)
Description
| Product Name | Quantity | Price (USD) | |
|---|---|---|---|
| Easy ID™ | 5 units at standard protocol | 355 | |
| Contents | |||
| *DMAP | DMAP-Lys(DMAP)-Lys(DMAP)-[Pro]6-Gly-MDA-N-maleimido | MW: 1521.8 | 0.1 mg |
| **Acyl donor | 5(6)FAM-[g-Abu]-S-Ph | MW: 553.1 | 0.1 mg |
Protocol
◆Maleimide DMAP modification protocol
- Dissolve the desired protein, antibody, etc. in an appropriate buffer (pH 7 to 8 recommended) such as PBS.
Finally prepare 50 μM, 300 μl of protein solution ①.
(The protein solution can be 50 μl – 500 μl depending on the actual experiment scale.) - Add 100 μl DMSO to this reagent to make 5 mM reagent stock ②.
(It seems it would be better to add 50 μl or 100 μl DMSO in one bottle and make 5 mM stock.) - Add 3 μl of equimolar reagent (DMSO stock solution ② above) to the protein of ① above and thoroughly mix the reaction solution by pipetting.
- Incubate at room temperature for 30 minutes to 60 minutes.
(It is better to follow the reaction with MALDI MS. If the reaction does not proceed 100%, add 1 to 2 equivalent of reagent stock.) - Purify the modified protein by gel filtration or the like.
Tips
During antigen labeling on the cell, the protein (antibody)-DMAP conjugate prepared above is made to have a final concentration of 1 μM.
Also, add Acyl donor to the cells (in the medium) to a final concentration of 8 μM. (See Hayashi et al., JACS (2015).)
Caution
- When the modification reaction does not proceed, it is recommended to reduce the protein of interest by DTT or TCEP. After reduction, remove surplus reducing agent.
- Incubation under nitrogen atmosphere is recommended to prevent re-oxidation of free cysteine.
◆Maleimide DMAP Modification Protocol (Reduced Version)
- Dissolve the desired protein, antibody, etc. in an appropriate buffer (pH 7 to 8 recommended) such as PBS.
Finally make 50 μM, 300 μl protein solution.
(Protein solution can be 50 ul – 500 ul depending on experiment scale.) - Add 2-5 equivalents of DTT or TCEP to the protein.
(You can use 5 mM reducing agent solution in the same buffer as protein.) - Incubate at 4 °C for 1 hour.
- In case of DTT exclude extra DTT with spin column.
(For TCEP go to the next step.) - Add 100 μl DMSO to this reagent to make 5 mM reagent stock ②.
(It seems it is better to add 50 μl or 100 μl DMSO in one bottle and make 5 mM stock.) - Add 3 μl of equimolar reagent (DMSO stock solution ② above) to the reduced protein and mix the reaction solution thoroughly by pipetting.
- Incubate at room temperature for 30 minutes to 60 minutes.
(It is better to follow the reaction with MALDI MS. If the reaction does not proceed 100%, add 1 to 2 equivalent of additional reagent stock.) - Purify the modified protein by gel filtration or the like.
Caution
To prevent re-oxidation of free cysteine, incubation under nitrogen atmosphere is recommended.
Inquiry / Order
Please send e-mail to the address s.anahara biologica.co.jp .
biologica.co.jp .
